Overview: my research involves the development and application of a range of high-dimensional single-cell cytometry/imaging technologies and assays, as well as computational analysis approaches, to map dynamic immune responses over time, space, and disease. Previously with the King Lab during my PhD, and currently with the Sydney Cytometry Facility and the collaborative Immune Dynamics Team, my team and I collaboratively apply these approaches to the study of immunology and infectious disease, including emerging pathogens such as SARS-CoV-2 and COVID-19, Zika virus encephalitis, and West Nile virus encephalitis. We then explore how these datasets, technologies, and analysis methdologies may contribute to, or benefit from, efforts such as the Human Cell Atlas (HCA).
This work is summarised in the following four sections (below):
- High-dimensional cytometry and imaging technologies
- Computational analysis approaches
- Application to disease (incl. COVID-19)
- Engagement in the Human Cell Atlas (HCA) and other communities
An overview of this work can be found in this Oz Single Cell webinar 2020 on computational biology and this Fluidigm webinar 2020, featured on the ‘COVID-19 resources’ page.
High-dimensional cytometry and imaging technologies

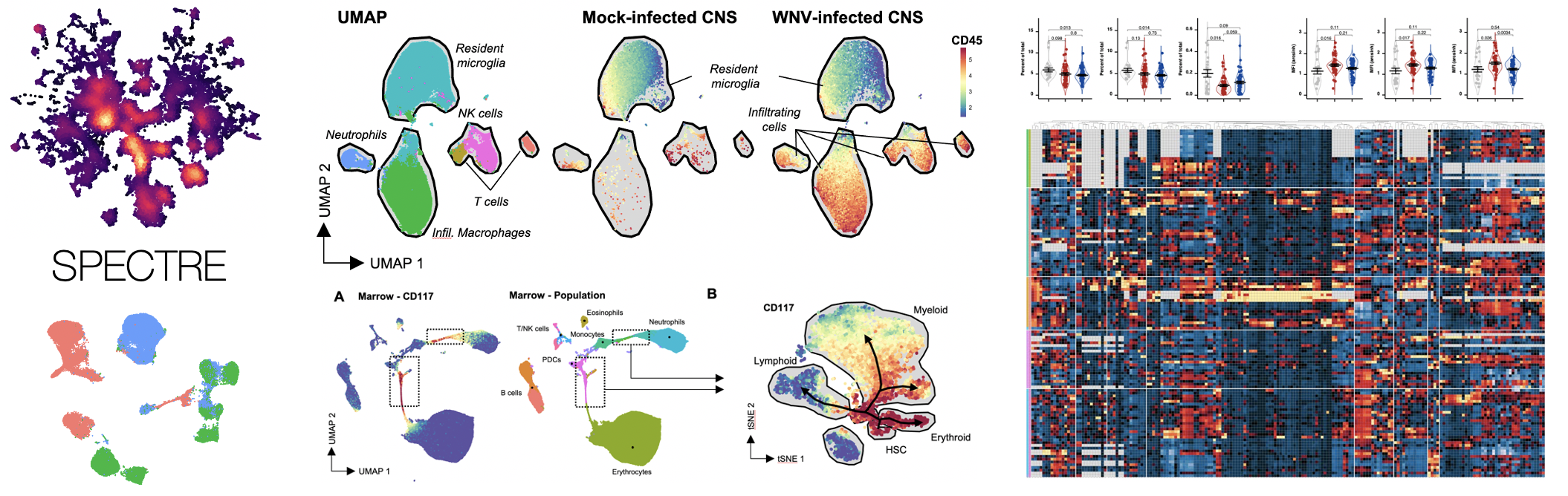
Critical to investigating complex immune responses to disease is the capacity to measure cells at the single-cell level, examining multiple features of the cell simultaneously. To address this, we develop and apply a variety of high-dimensional cytometry technologies and assays. We have developed novel technology platforms, such as our world first 10-laser ‘LSR-X’ platform developed with BD, and one of the first published 25 colour flow cytometry assays (Ashhurst et al. 2017). We also developed the first assays to be used on Australia’s first mass cytometer (CyTOF) (Ashhurst et al. 2019), as well as Australia’s first Imaging Mass Cytometer (IMC, SpectreMAP Github). We have recently published a book (HM McGuire, TM Ashhurst (eds). 2019), featuring a collecting of up-to-date and cutting-edge protocols in mass cytometry, which is now a standard resource in the field. Most recently, we have implemented high-dimensional spectral cytometry platforms and panels, and performed comprehensive evaluations of both conventional and spectral cytometry (Niewold*, Ashhurst* et al. 2020).
Computational analysis approaches

Following a seed funding grant from the Marie Bashir Institute for Infectious Diseases and Biosecurity (“Mapping dynamic immunity: next-generation computational approaches to track the evolution of immune responses in West Nile virus and Zika virus encephalitis”), we established the ‘Immune Dynamics’ team, a collaborative group with a focus on the development of novel computational analysis tools to address challenges in high-dimensional analysis.
My team and I have developed a number of computational analysis approaches, including SPECTRE, an R package that enables comprehensive end-to-end integration and analysis of high-dimensional cytometry data from different batches or experiments (Ashhurst*, Marsh-Wakefield*, Putri* et al. bioRxiv. 2020). To facilitate the analysis of time-series data, we have also developed a number of time-series clustering algorithms, including ChronoClust (Putri et al. 2019) and TrackSOM, which we recently applied to the study of COVID-19 (Koutsakos et al. 2021). We have also developed novel multi-perspective methods for the evaluation of clustering algorithms (Putri et al. 2021). Most recently, we have adapted the Spectre package to facilitate spatial analysis of IMC data (SpectreMAP Github). Our ongoing work is focused on the development of better data integration approaches and spatial analysis tools.
Application to disease

We apply our high-dimensional technologies and analysis approaches to study inflammation and infectious disease. A particular focus has been the study of immunopathology, where the immune response to infection may be a significant driver of disease.
SARS-CoV-2 and COVID-19
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has caused >101 million infections and 2.18 million deaths worldwide (as of January 28, 2021). Infection with SARS-CoV-2 results in a spectrum of clinical presentations, called coronavirus disease 2019 (COVID-19), ranging from asymptomatic to fatal disease. Through a collaboration with the Doherty Institute and the University of Melbourne, we comprehensively profiled the immune response to SARS-CoV-2 in a cohort of COVID-19 patients. Our work demonstrated that patients with severe disease exhibited an excessive and hyper-activated immune response (Koutsakos et al. 2021). To study the kinetics of this immune response, we also developed and applied a novel time-series analysis approach called TrackSOM.
- This study has been featured on Doherty Instite News, Medical Xpress, the Herald Sun, and Ticker.
- Our webinar from 2020 (Mapping Immunity Across Time, Space and Disease State) is featured on the Fluidigim ‘COVID-19 resources’ page.
- Our mass cytometry protocols have been cited in a number of COVID-19 studies, including Rodriguez et al. 2020 and Koutsakos et al. 2021.
- Panel design and analysis protocols that may be helpful in COVID-19 research can be found on the resources page.
Zika and West Nile virus encephalitis and immune modulation
In mouse models of viral encephalitis, our work has demonstrated that inflammatory monocyte-derived macrophages which infiltrate into the brain contribute significantly to disease in Zika virus encephalitis (Hayashida et al. 2019) and West Nile virus encephalitis (Getts et al. 2012), and that immune-modulatory therapies targeting inflammatory monocytes may inhibit this process (Getts et al. 2014). This work was featured on ABC News and The Guardian. Our most recent work has demonstrated that viral encephalitis drives an inflammatory mobilisation of the haematopoietic system in the bone marrow, resulting in the generation of these pathogenic monocytes (Ashhurst et al 2019, Ashhurst 2020, Fluidigm webinar).
Immunology
Through various collaborations, we have applied our approaches in a number of areas, such as IRF8 depletion (Terry et al. 2015), T cell exhaustion in LCMV infection (Huber et al. 2017), VZV infection of NK cells (Campbell et al. 2018), antibody-mediated cell subset depletion (Jung et al. 2018), interferon-mediated lethality in LCMV infection (Jung et al. 2020), B cell responses in multiple sclerosis (Marsh-Wakefield et al. 2020), and intrapulmonary vaccination strategies (Ferrell et al. 2021).
Engagement in the Human Cell Atlas and other communities

We are actively involved in the Human Cell Atlas (HCA) community, including attendance at HCA general meetings in Hinxton, UK (2018), and Tokyo, Japan (2019), seeking to help define the contribution of high-dimensional cytometry and imaging technologies to the HCA objectives (Czechowska et al. 2018, WS06: ‘Cytometry in the Era of the Human Cell Atlas’). We also presented on Spectre, one of our computational tools, at the HCA Asia meeting (virtual) in 2020.

We have taken a key role in two roadshows hosted by the Australasian Cytometry Society (ACS): Polychromatic Flow Cytometry in 2016 (Dr. Pratip Chattopadhyay and Thomas Ashhurst), and High-Dimensional Cytometry Analysis in 2020 (Dr. Thomas Ashhurst, Dr. Felix Marsh-Wakefield, and Givanna Putri).
